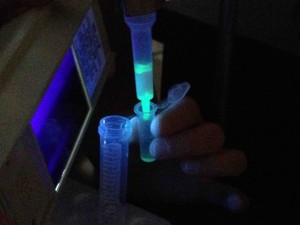
In the “Bioreactor Cell Culture and Protein Recovery” course at Laney College, my students recently transformed bacteria with two different sources of DNA to create green and blue glowing bacteria. Here are the before and after shots:

IPTG induced bacteria under ambient light (left) and UV light (right).
Why did we do this experiment? For one, it looks really cool when you put a gene from a jellyfish into a bacteria and then the protein makes it glow under UV light. But the real reason was to compare protein expression systems in E. coli, a commonly used “chassis” in biotechnology, biomanufacturing, and the new field of synthetic biology.
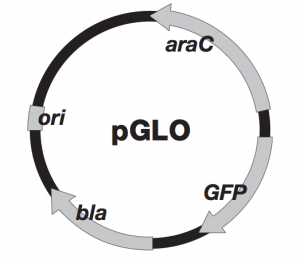
In past experiments, we have used a pGLO plasmid in which the gene for the glowing protein, green fluorescent protein (GFP), is induced (or “turned on”) by the presence of a sugar called arabinose. The plasmid also contains resistance to ampicillin, an antibiotic that inhibits the cell wall formation in gram-negative bacteria like E. coli. Here is a map of the arabinose induced pGLO plasmid:
How the arabinose induced plasmid works:
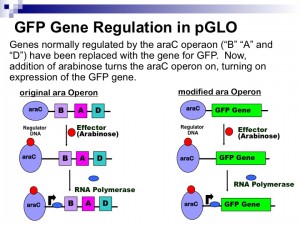
Arabinose is a simple plant sugar and is used as food by bacteria like E. coli. The bacterial genes for arabinose digestion are not expressed unless arabinose is present. No need to waste all that energy on nothing, right? When arabinose is present, however, these genes get turned on and arabinose is broken down. When arabinose runs out, the genes turn off. Three structural genes (araB, araA, araD or BAD) encode for enzymes used in the digestion of arabinose in bacterial cells. Another gene called araC produces a protein that regulates the expression of BAD. The araC protein also acts to negatively control its own synthesis. In other words, the araC protein binds to its own operator and prevents more araC transcription. The presence of both the araC protein and arabinose cause the expression of BAD. Arabinose binds to the araC protein and causes a change in its shape, which then allows both to bind to the regulatory region known as the activator site. The binding of the araC protein-arabinose complex to the DNA activator site allows the binding of RNA polymerase to the promoter region upstream of BAD. These three genes can now be transcribed and the resulting RNA can be translated into protein. Thus, araC negatively controls its own expression and positively controls BAD. If you replace BAD with the GFP gene, you can then positively regulate the production of GFP using arabinose. Here is a summary figure of the arabinose operon:
This expression system has given us excellent results. We follow up the GFP production with a purification step, which looks like this:
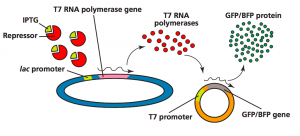
The protein that was produced in the first picture above (green and blue glowing bacteria) used a different expression system involving the lac operon. The lac operon is induced by IPTG, an analog or similarly shaped molecule to lactose. Here is a diagram showing how the IPTG induction works:
How the IPTG induced plasmid works:
Lactose is another sugar that can be used as an energy source by bacteria like E. coli. Bacteria have three genes involved in metabolizing lactose when it is the only carbon source present (bacteria will use glucose first if both lactose and glucose are present): lacZ, lacY, and lacA. These three genes are considered inducible because they will “turn on” when lactose is present. Another gene, lacI (that is the letter “i” for “inducible”), produces a repressor protein. When lactose is absent, the repressor binds to the operator which overlaps the promoter region and prevents transcription of the XYA genes. When lactose is present it binds to the repressor protein and prevents it from binding to the DNA operator. Then, RNA polymerase can transcribe the XYA genes and lactose will be metabolized. This is another example of negative control. Here is a diagram of how the lac operon works:
- downstream gene.
Comparing the two fluorescent protein expression systems:
As mentioned above, the lac operon can be induced by IPTG, which does not get broken down by the proteins encoded by the normal lac operon. Arabinose gets eaten as food by bacteria so the induction concentrations are typically much higher than with IPTG. Arabinose induction is done at a 2-6 g/L final concentration, whereas IPTG induction is done at 0.12-0.24 g/L final concentration.
There are advantages to using arabinose as well. Arabinose induction is titratable, meaning that the higher the concentration of arabinose, the stronger the expression. Therefore, protein expression can be adjusted which may be important if the protein has toxicity or solubility problems. Another advantage is that arabinose is also less expensive than IPTG.
One thing I noticed with the IPTG induced green and blue fluorescent protein fermentations was that there appeared to be a little glowing before IPTG induction. It could be an indication of background fluorescent protein production that can happen with this system. Another E. coli strain called BL21(DE3)pLysS has a T7 lysozyme engineered into it. The T7 lysozyme decreases the background expression level of genes behind the T7 promoter but does not block target protein expression following induction by IPTG. Perhaps this would be something we would look at using if background expression was becoming a problem.
Both systems of fluorescent protein expression have their advantages and disadvantages. The real test will come later when we scale up our production volumes from 100 milliliters (mL)(shown above) to 1 or 3 liters (L). Also, we will want to test both systems in downstream processing performance. Which will give the higher protein titers? Will insoluble protein inclusion bodies be a problem with the higher expression levels? We will find out later this semester!